
Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration.Molecular docking is a powerful technique that helps uncover the structural and energetic bases of the interaction between macromolecules and substrates, endogenous and exogenous ligands, and inhibitors. Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading.

The PyMOL Molecular Graphics System, Version 1.8.
Seeliger D, de Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Sandeep G, Nagasree KP, Hanisha M, Kumar MMK (2011) AUDocker LE: a GUI for virtual screening with AUTODOCK Vina. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera-a visualization system for exploratory research and analysis. Nissink JW (2009) Simple size-independent measure of ligand efficiency. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity.
#JMOL SIMULATION HOW TO#
Maxmen A (2016) Busting the billion-dollar myth: how to slash the cost of drug development. Lill MA, Danielson ML (2011) Computer-aided drug design platform using PyMOL. Jiang X, Kumar K, Hu X, Wallqvist A, Reifman J (2008) DOVIS 2.0: an efficient and easy to use parallel virtual screening tool based on AutoDock 4.0.
#JMOL SIMULATION FREE#
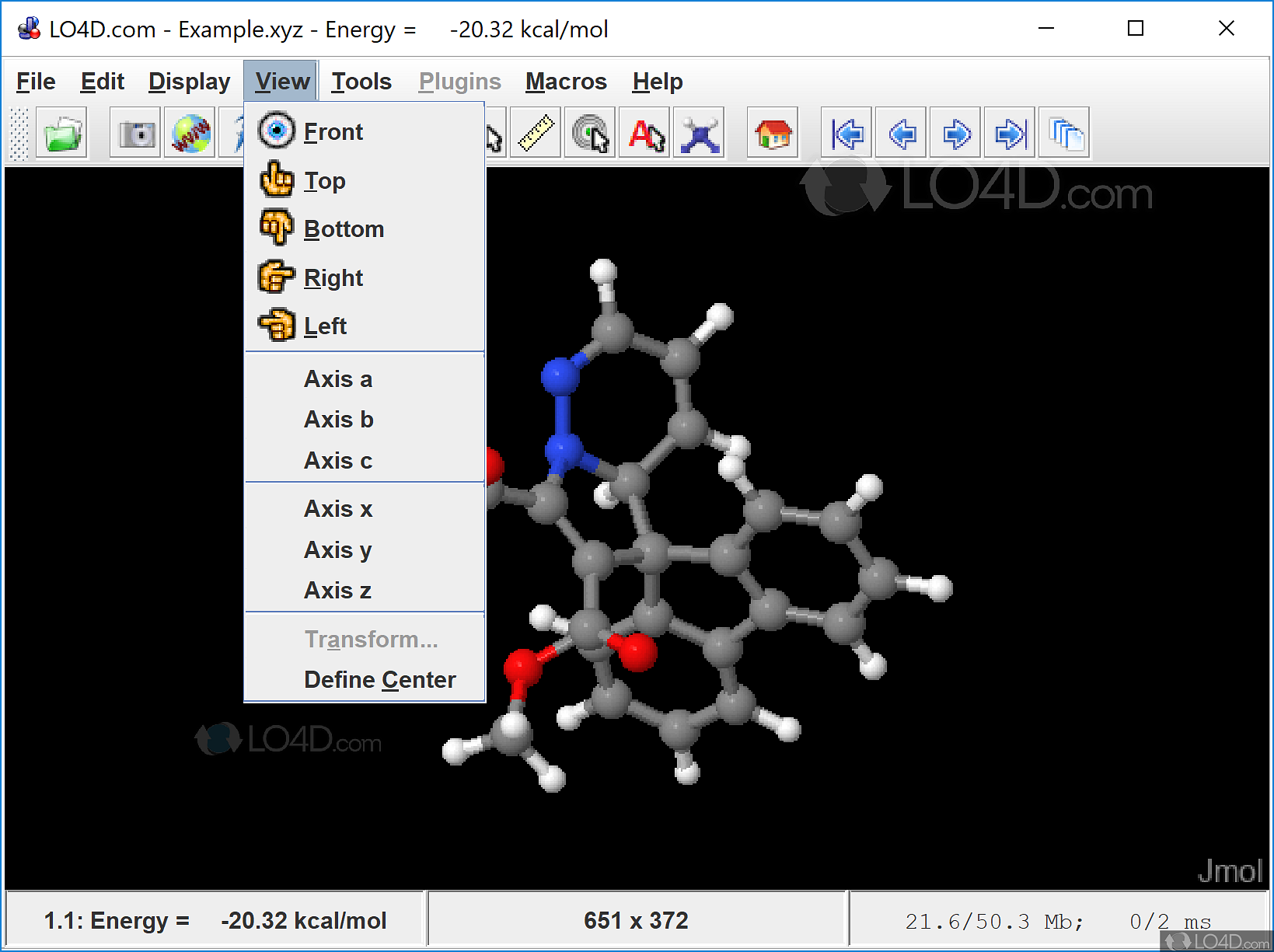
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) ZINC: a free tool to discover chemistry for biology. Hanson RM (2010) Jmol-a paradigm shift in crystallographic visualization. Gaudreault F, Morency LP, Najmanovich RJ (2015) NRGsuite: a PyMOL plugin to perform docking simulations in real time using FlexAID. Furthermore, other databases of compounds such as ZINC, available also in AutoDock format, can be readily and easily plugged in. The application comes with the DrugBank set of more than 1400 ready-to-dock, FDA-approved drugs, to facilitate virtual screening and drug repurposing initiatives. DockingApp sports an intuitive graphical user interface which greatly facilitates both the input phase and the analysis of the results, which can be visualized in graphical form using the embedded JMol applet. Along this path, here is described the implementation of DockingApp, a freely available, extremely user-friendly, platform-independent application for performing docking simulations and virtual screening tasks using AutoDock Vina.
#JMOL SIMULATION FULL#
The need to promote community-driven drug development efforts, especially as far as neglected diseases are concerned, calls for user-friendly tools to allow non-expert users to exploit the full potential of molecular docking. Moreover, this technique plays a pivotal role in accelerating the screening of large libraries of compounds for drug development purposes. Molecular docking is a powerful technique that helps uncover the structural and energetic bases of the interaction between macromolecules and substrates, endogenous and exogenous ligands, and inhibitors.


 0 kommentar(er)
0 kommentar(er)
